How to make data points worthwhile?
Over the years as we worked with more and more clients, we encountered training data in all shapes and sizes. It quickly became evident that just having a lot of raw data points doesn’t automatically result in a good model - it matters what you measure.
The team began developing intuitions about which types of training data work best. For example, drug perturbations seemed about 10 times more impactful than the same number of CRISPR data points. Post-treatment RNASeq data (measuring the whole transcriptome after treatment rather than just cell viability) gave better models, but definitely not 20.000 times better1. Similarly, single cell measurements are better than bulk measurements, but not 10.000 times better2.
I asked to have these intuitions quantified.
Why is CRISPR worse? Is the technology inherently less robust?
It matters what you perturb
Turns out the problem was that cells are pretty robust so most single gene KOs don’t really do anything. Drugs, on the other hand are specifically designed to elicit a response and usually bind to multiple proteins especially at higher concentrations.
Therefore, in the CRISPR training we were showing a lot of differently looking RNASeq data to the model, but most of those amounted to no actual change in cell behavior, essentially teaching noise to the model. No wonder these models performed worse.
Compressing training data
In this case, is it true that only perturbations that cause meaningful changes carry most of the information in the training? Could we “compress” the training data by only using the meaningful data points?
In order to do that, the first question we had to answer was what “meaningful change” means.
Different teams came up with different definitions.
“Horizons” team devised the idea that a sample is significant if the RNASeq pattern is significantly different in any pathway. This gives a compression factor of ~6x, so 1500-2000 points from every 10000 data points in LINCS.
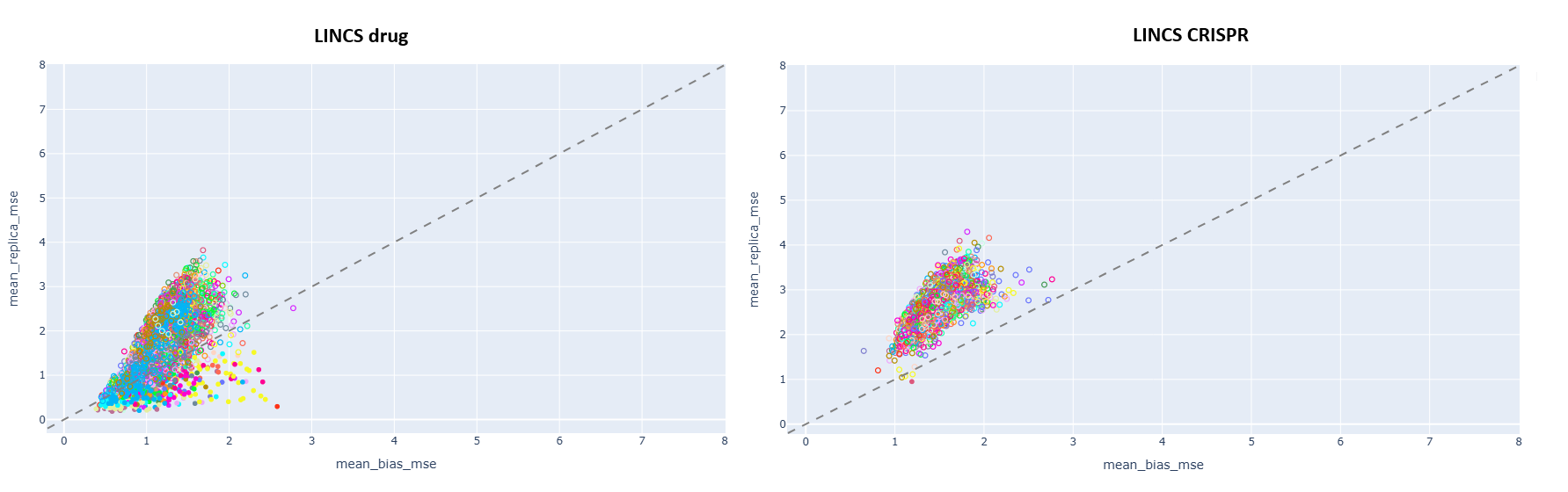
“Skunkworks” took the RNA pattern as a whole, hypothesizing that those RNASeq patterns are useful where the replicates are closer to each other than to the bias (average RNASeq value over many perturbations). This is the data you can see on the first figure above.
Surprisingly, samples where this is true are very rare, yielding a compression factor of 30-50x (2-300 significant samples from 10000)
So how well do these compressed datasets work in an actual training?
Here’s how.

Disclaimer: this was just a quick investigation, not a fully robust analysis.
What the results appear to suggest though, is that it is at least possible to find small subsets of data points carrying most of the useful information in a training
(this is shown by the performance of the “filter” column being equal or better than the baseline “full” dataset performance while at the same time datasets trained on the inverse subsets (“inverted”) missing these key data points performed significantly worse.)
Given how little perturbed RNASeq data is out there to train from, there is no need to speed up trainings with data compression yet. We still do some subsetting to remove the noise, but otherwise generally try to use as much info as we can.
A better way to generate wet-lab data
The real value of this compression becomes apparent when generating wet-lab data. Wouldn’t it be wonderful if there was a way to only generate the important data points? To get 6-30x the value for your experimental dollars?
The problem is that we only know whether a perturbation did anything after performing the experiment itself.
But what if there was a way to guess?
Maybe if you had something like, I don’t know, a Simulated Cell?😊
Let’s run some numbers, how much could such a predictive model help?
Based on the previous tests, the prevalence of meaningful events in a randomly generated data set is somewhere around 10% .
Now suppose you have a predictive model where you both have a 70% chance to successfully predict these meaningful events, but you also have a 30% chance to have false negatives (70% sensitivity). This results in the following contingency table.
So, generating 100 “smart” data points yields 70 valuable training samples compared to the 10 we would have originally gotten using random generation:a 7x compression rate.
The catch is that there are important points you will always discard due to the false negatives of the simulator, 3.3% of the significant data points in this case.
It’s a tradeoff I’m willing to take.
In real life, our actual performance varies depending on how hard the problem is (see the previous benchmarking post)
but it’s around 60-70% for the hardest specific subsets with a lot of outliers (resulting in 6-7x compression rate just as shown in the table above) and around 95% on the full dataset, yielding a 20x effective data compression rate.
AI in biology can be done today if you use data wisely
We believe that for many practical use-cases in drug discovery, AI for complex biology is already possible, today.
However, to make your platform viable in current industry practice, you need to use your data wisely. The reality is that you can’t go around asking for hundreds of thousands of training data points to fine-tune every new project you get, because very few pharma projects generate that kind of data. At the same time, few drug discovery teams out there would be able or willing to accept the data generation “fit-out phase” for their indications of interest to take more than a year and cost many millions of dollars. It’s just not sustainable.
But if you can provide useful results, quickly, from datasets of a size that pharma companies can actually generate in realistic time (and cost) and keep doing that, it will add up over time.
In the end, probably no single AI platform will be able to match the simultaneous data generation capability of all pharma companies rushing to get their next drug to market. So we think our best bet now is to tap into that stream.
That necessitates a platform that can provide partners value now. Which is why we’ve always had dedicated teams working on making our production models actionable and easy-to-use rather than only focusing on the next model generation.
Data kindly lifted from the work performed by Laszlo Mero, Milan Sztilkovics, Imre Gaspar and Valer Kaszas - thank you for your work! Additional thanks for the review & edits to Balint Paholcsek, Szabolcs Nagy, Imre Gaspar and Bence Szalai.
RNASeq generates one data point per gene in each sample.
Single-cell methods measure somewhere between 1000 to 10000 cells per sample.



