Pretraining virtual cells is useless
the way we do it now
UPDATE: this was extended into a poster at scVerse 2025, you can check it out here.
Clickbaiting aside, theoretically, pretraining is still a good idea:
Let’s try to make use of the troves of unlabeled, pre-treatment single-cell RNASeq data to teach the system biological fundamentals. Then the actual training on the targeted, expensive, and scarce data doesn’t have to start from ground zero, so maybe it goes farther.
Before we dive in and talk about the results, a quick primer on pretraining.
What are we pretraining?
What does the AI really learn during pretraining? There is no shortage of fancy names like “language of life” or “fundamental biology” (yes, I am guilty too). But deep inside, it’s just correlations of gene expression. Something like this: if gene A is highly expressed and gene B is not expressed, then most likely gene C is highly expressed as well.
Some articles mention learning gene regulatory networks (GRNs). It’s the same thing. If you take these complex correlations, simplify them to 1-to-1 rules and chain them together, what you get is a GRN.
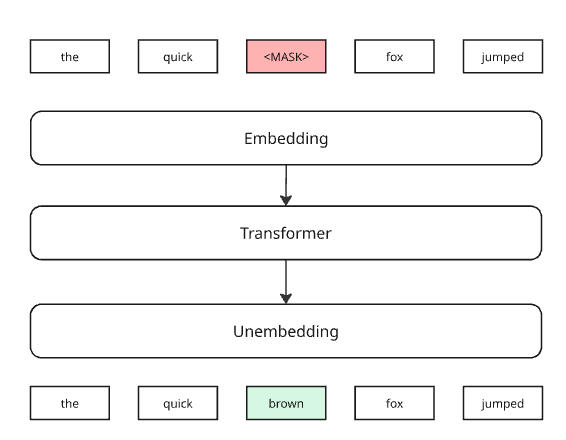
The way most bioGPTs go about this pretraining is that we mask some of the expression values, and ask the system to predict them.
Here’s how it would look in an LLM:
It’s like asking the system to correct some broken English. There’s a word that’s clearly wrong there, what the author could have intended?
Proof of the pudding
So one of our research teams went in, and did the same thing - masked out 15% of the gene expressions in all samples. They then ran a pre-training on scRNASeq data from CELLxGENE, 30M cells (filtering out 10% of the original 33M samples not coming from the 10x Genomics platform).
It worked perfectly. The predictions had a 76% correlation with the ground truth, the same as experimental replicates. Great!
Then came fine-tuning. Ultimately we want to understand how drugs affect the cells. We want to be able to predict how their transcriptomic landscape looks after treatment. So we added some perturbation data.

Not much difference. If anything, fine-tuning made the training slightly worse. So to understand drug response1, pretraining was not really helpful. But maybe we were doing it wrong. How much information is really there in untreated RNA data?
Less information than you think
So the team went back to work. The interesting moment came with a set of experiments where they started increasing the ratio of masked genes.

Wait, what? We are at the experimental replicate level of predictivity even with all genes masked out? What does the system even base its predictions on?
Turns out the remaining information with all genes’ expressions masked out is which genes were even expressed in the cell to start with. Single-cell RNASeq differs from bulk in that it captures relatively little RNA from each individual cell, so what you see is just the most abundant genes rather than the the full transcriptome.
The issue isn’t that more information from RNASeq doesn’t help - that can be on us. The problem is that this piece of information is enough to get to experimental-level performance - ie. that’s all the information there is about single cells!
It seems that in pre-treatment cells the genes detected by scRNASeq are heavily correlated. Probably there aren’t that many major cell types to start with, and single-cell technology is not yet good enough to reliably detect smaller changes in individual cells.
Which leaves us with three possible consequences:
either the gene regulatory information in the data is not good enough
or we’re not extracting data the right way (there is additional information in how cells associate which we’re not using)
or healthy gene regulation is just not the right kind of information we need to understand drug response.
Thanks to our great Skunkworks Infinity team — Gerold Csendes, Gema Sanz, Bence Czako and Bence Szalai for working this out!
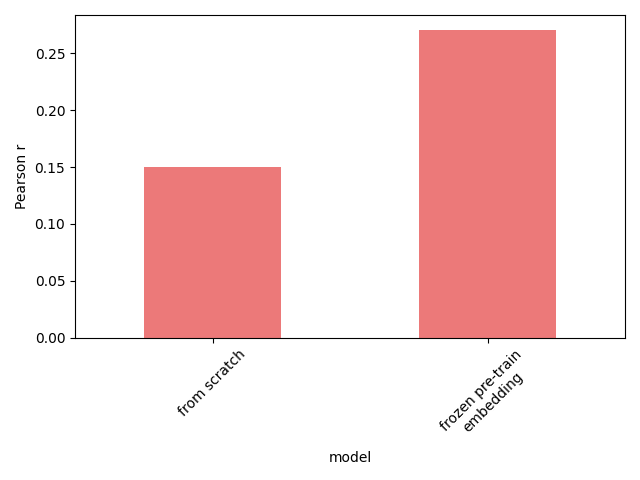
We could design a very specific task where the missing information is gene regulation, and pre-training does help - predicting unknown gene expression values post-perturbation given some of the post-treatment expression values from the same perturbation. The figure below shows the effect of pre-training. Whether this is useful in any practical application is anyone’s guess.